Abstract
We have designed a multiplex ligation-dependent probe amplification (MLPA) assay to simultaneously screen all 79 DMD gene exons for deletions and duplications in Duchenne and Becker muscular dystrophy (DMD/BMD) patients. We validated the assay by screening 123 unrelated patients from Serbia and Montenegro already screened using multiplex PCR. MLPA screening confirmed the presence of all previously detected deletions. In addition, we detected seven new deletions, nine duplications, one point mutation, and we were able to precisely determine the extent of all rearrangements. To facilitate MLPA-based screening in laboratories lacking specific equipment, we designed the assay such that it can also be performed using agarose gel analysis and ethidium bromide staining. The MLPA assay as described provides a simple and cheap method for deletion and duplication screening in DMD/BMD patients. The assay outperforms the Beggs and Chamberlain multiplex-PCR test, and should be considered as the method of choice for an initial DNA analysis of DMD/BMD patients.
Similar content being viewed by others
Introduction
The Duchenne muscular dystrophy (DMD) gene is the largest known gene, spanning ∼2.4 Mb of genomic sequence on Xp21.1 Mutations in the gene cause DMD, the most commonly inherited neuromuscular disorder, and Becker muscular dystrophy (BMD), the milder allelic form of the disease. The mutation spectrum within the gene is unusual in that deletions of one or more exons are found in ∼65% of cases.2, 3, 4 Deletions are known to cluster in two hotspot regions,4, 5 and as DMD is X-linked, these can be easily detected in males using simple PCR reactions. Two multiplex PCR kits of nine exons each were developed that together detect ∼95% of all deletions.6, 7
Although a significant proportion (5–8%) of duplications was reported in DMD and BMD early on,3, 8 most laboratories do not systematically screen for these rearrangements. This is because duplication analysis and the determination of carrier status require a quantitative method of analysis, which is more laborious and technically demanding. Until recently, the most commonly applied methods have been Southern blotting,2, 3 including pulsed-field gel electrophoresis, and quantitative multiplex PCR.9, 10
We have previously reported the use of multiplex amplifiable probe hybridization (MAPH) for screening all 79 exons of the DMD gene for deletions and duplications in two reactions.11 Recently a similar technique was developed, multiplex ligation-dependent probe amplification (MLPA).12 The ease of use made MLPA a popular technique that has been widely applied. The disadvantage of MLPA compared to MAPH is the more laborious and costly design of a good probe set. To overcome this limitation we designed a DMD gene MLPA set, the use of which has recently been reported.13, 14 In this study, we describe its performance in screening for deletion and duplication mutations in a series of DMD and BMD patients from Serbia and Montenegro.
Methods
Patient samples
A group of 133 patients (98 DMD and 35 BMD) referred to the Laboratory of Medical Genetics, Mother and Child Health Institute of Serbia, Belgrade, for molecular analysis of the DMD gene were screened. Genomic DNA was extracted from leukocytes by a salting out procedure.15
Multiplex PCR deletion screening
For multiplex PCR screening, 18 exons (3, 4, 6, 8, 13, 19, 30, 42, 43, 44, 45, 47, 48, 50, 51, 52, 53 and 60) and the muscle promoter of the DMD gene were tested,16 although not all samples were tested for exons 30 and 53. Additional primers were used to further delineate the 5′ or 3′ exonic extent of some deletions, that is, the primers for exons 12 and 46,6 16 and 41,17 and 49.7 The PCR products were separated on a 2% agarose gel.
Multiplex ligation-dependent probe amplification
The MLPA reactions for probe sets P034 and P035 (MRC-Holland, Amsterdam, The Netherlands) were performed essentially as described,12 except that the PCR reaction was in a final volume of 25 μl, and 33–35 cycles were performed. Initial analysis was performed by eye, with a deletion in a male patient defined as no visible peak at the expected position. For the remaining samples, the height of each exon-specific peak was divided by the two nearest control peaks. The median ratio across all samples for each peak was calculated and used as a reference for one copy (corresponding to the single X-chromosome in males). Although a deletion would be expected to lead to the total absence of a peak and hence a ratio of zero, any normalized ratio below 0.5 was considered a possible deletion. This takes into account the possibility of a change in sequence around the ligation site of the MLPA half-probes, which may disturb the ligation reaction sufficiently to reduce the PCR yield of the probe. Unless the result matched precisely with what was previously found, all apparent single exon deletions were confirmed by independent MAPH analysis, as previously described,11 or by PCR amplification and sequencing.
A duplication would be expected to give a normalized ratio of 2. We set the threshold at 1.5, although to minimize false-negative results we retested any probe with a normalized ratio above 1.4. Any cases involving a single exon were retested in a duplicate MLPA reaction.
The MLPA reactions using probe sets P071–P078 (MRC-Holland, Amsterdam, The Netherlands) were performed as described,12 except that the PCR amplification was for 35 cycles, and 15 μl unlabeled PCR product was separated on a 3% agarose gel. Analysis was performed visually.
Results
Multiplex PCR
A total of 133 DNA samples from both DMD and BMD patients were analyzed. Of these, 123 were unrelated cases, and the results described are based on these only. The samples had been screened for deletions by multiplex PCR, resulting in the detection of 71 deletions.
Multiplex ligation-dependent probe amplification
MLPA analysis of the same samples revealed 88 rearrangements: 78 deletions, nine duplications and one point mutation. The average ratio of the duplicated exons was 1.88 (range, 1.44–2.46), and the average ratio of triplicated exons was 2.73 (range, 2.52–3.12). All new mutations detected are listed in Table 1.
Only one deletion, of exon 15, could not be confirmed by another technique. Sequence analysis revealed a nonsense mutation (NM_004006.1; c.1729 G>T, p.Glu577X). The altered nucleotide was located precisely at the ligation site of the two half-probes, disturbing the ligation sufficiently to appear as a deletion.
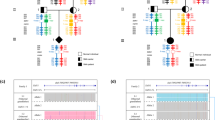
An unusual rearrangement was found in two brothers, apparently composed of 2 non-contiguous duplications and a partial triplication. The duplications cover exons 52–55 and 63–67, while the triplication covers exons 68–79 (Figure 1). The full extent of the rearrangement after the last exon was not determined.
Analysis using the P034 and P035 probe sets as described above requires the availability of an electrophoresis system able to detect labeled probes (a sequencing system). Such equipment will not be available in every laboratory and therefore we analyzed the MLPA products using simpler methodology, that is, agarose gel electrophoresis and ethidium bromide staining. To facilitate this analysis the probe sets were divided into eight sets (P071–P078), with fragment sizes differing by at least 20 bp ensuring easy discrimination after electrophoresis (Figure 2a). A series of samples with known deletions were screened with several of the probe sets. In all cases the boundaries perfectly matched those determined previously (Figure 2b). To test the limits of the system we analyzed a sample from a male patient with a duplication of exons 58–63. Comparing band intensities between the patient and the control sample clearly showed that the exon 59 band gave a relatively stronger signal in the patient sample (Figure 2c).
Images using the probe sets developed for agarose gel analysis. (a) The eight probe sets (P071–P078), together covering every exon of the DMD gene. Each set includes at least one control probe. (b) Results obtained from samples with a deletion of one or more exon. Lanes 1 and 2 show the products derived from patients with previously identified mutations, and lane 3 is from a control sample. The missing bands in lanes 1 and 2 correspond with the deleted exons. (c) An example of a duplication visible by agarose gel electrophoresis. Lane 1 is from a patient with a previously identified duplication, lane 2 is from a female control. The band indicated by an arrow is significantly more intense in lane 1 than lane 2, corresponding with the duplication of exon 59. The control band is also stronger in the patient compared with the control, this is due to the sex mismatch of the two samples.
Discussion
We describe the use of MLPA for the detection of exonic rearrangements in the DMD gene. Compared to analyzing the deletion hotspot regions by multiplex PCR, the advantages of MLPA-based screening of all exons are obvious. First, 13% more mutations were detected; 7% duplications and 6% deletions. In addition, the full extent of 20 deletions was not determined prior to MLPA analysis. This knowledge is critical when predicting the progression of the disease; in ∼90% of cases a reading-frame-disrupting mutation will lead to the lethal Duchenne phenotype, whereas mutations that maintain the open reading frame in majority of cases lead to the milder Becker-like phenotype.18 This information will also be of importance for future gene therapy strategies, for example, those targeted at the skipping of specific exons of the gene.19, 20
For laboratories without access to sequencing systems capable of detecting fluorescent labels, we designed probe sets allowing agarose gel electrophoresis. Since the resolution of agarose gels is lower than that of polyacrylamide gels or capillary electrophoresis, fewer probes can be combined per reaction, meaning that eight lanes are needed to cover all 79 DMD exons. Since DMD is an X-linked disease, deletion analysis in males is simple and the complete extent of the deletion is immediately apparent. Our results indicate that it seems possible to detect duplications on the agarose gel as well, in particular, when a series of exons is duplicated.
The DMD MLPA test provides a simple and cheap DNA-based test for deletion/duplication screening that can be performed in any DNA laboratory. As its performance clearly outperforms the Beggs and Chamberlain multiplex-PCR test6, 7 it should be considered as the method of choice for a first DNA analysis of DMD/BMD patients. After a negative MLPA-test, the gene can be scanned for point mutations using RNA21, 22 or DNA-based tests.23, 24, 25
References
Den Dunnen JT, Grootscholten PM, Dauwerse JD et al: Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination. Hum Mol Genet 1992; 1: 19–28.
Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener CA, Kunkel LM : Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 1987; 50: 509–517.
Den Dunnen JT, Grootscholten PM, Bakker E et al: Topography of the DMD gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 1989; 45: 835–847.
Oudet C, Hanauer A, Clemens P, Caskey CT, Mandel JL : Two hot spots of recombination in the DMD-gene correlate with the deletion prone regions. Hum Mol Genet 1992; 1: 599–603.
Forrest SM, Cross GS, Speer A, Gardner-Medwin D, Burn J, Davies KE : Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature 1987; 329: 638–640.
Chamberlain JS, Gibbs RA, Ranier JE, Nga Nguyen PN, Caskey CT : Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 1988; 23: 11141–11156.
Beggs AH, Koenig M, Boyce FM, Kunkel LM : Detection of 98-percent DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 1990; 86: 45–48.
Hu X, Burghes AHM, Ray PN, Thompson MW, Murphy EG, Worton RG : Partial gene duplications in Duchenne and Becker muscular dystrophies. J Med Genet 1988; 25: 369–376.
Yau SC, Bobrow M, Mathew CG, Abbs SJ : Accurate diagnosis of carriers of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet 1996; 33: 550–558.
Ioannou P, Christopoulos G, Panayides K, Kleanthous M, Middleton L : Detection of Duchenne and Becker muscular dystrophy carriers by quantitative multiplex polymerase chain reaction analysis. Neurol 1992; 42: 1783–1790.
White S, Kalf M, Liu Q et al: Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet 2002; 71: 365–374.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G : Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002; 30: e57.
Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J : MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics 2005; 6: 29–35.
Schwartz M, Duno M : Improved molecular diagnosis of dystrophin gene mutations using the multiplex ligation-dependent probe amplification method. Genet Test 2004; 8: 361–367.
Miller SA, Dykes DD, PoleskY HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Abbs SJ, Yau SC, Mathew CG, Bobrow M : A convenient multiplex PCR system for the detection of dystrophin gene deletions: a comparative analysis with cDNA hybridisation shows mistypings by both methods. J Med Genet 1991; 28: 304–311.
Kunkel LM, Snyder JR, Beggs AH, Boyce FM, Feener CA : Searching for dystrophin gene deletions in patients with atypical presentations; in Lindsten J, Petterson U (eds): Etiology of Human Diseases at the DNA Level. New York: Raven Press, 1991, pp 51–60.
Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM : An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988; 2: 90–95.
Lu QL, Mann CJ, Lou F et al: Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med 2003; 9: 1009–1014.
Aartsma-Rus A, Janson AA, Kaman WE et al: Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet 2004; 74: 83–92.
Tuffery-Giraud S, Saquet C, Chambert S et al: The role of muscle biopsy in analysis of the dystrophin gene in Duchenne muscular dystrophy: experience of a national referral centre. Neuromuscul Disord 2004; 14: 650–658.
Roest PAM, Roberts RG, Van Der Tuijn AC, Heikoop JC, Van Ommen GJB, Den Dunnen JT : Protein truncation test (PTT) to rapidly screen the DMD-gene for translation-terminating mutations. Neuromusc Disord 1993; 3: 391–394.
Mendell JR, Buzin CH, Feng J et al: Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurol 2001; 57: 645–650.
Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB : Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet 2003; 72: 931–939.
Hofstra RM, Mulder IM, Vossen R et al: DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients. Hum Mutat 2004; 23: 57–66.
Acknowledgements
We thank the patients and their families for their cooperation and Dave van Heusden for expert technical assistance. Jan Schouten is director and shareholder of MRC-Holland, the producer of the MLPA kits described in this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lalic, T., Vossen, R., Coffa, J. et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet 13, 1231–1234 (2005). https://doi.org/10.1038/sj.ejhg.5201465
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201465
Keywords
This article is cited by
-
NGS-based targeted sequencing identified six novel variants in patients with Duchenne/Becker muscular dystrophy from southwestern China
BMC Medical Genomics (2023)
-
RNA-seq analysis, targeted long-read sequencing and in silico prediction to unravel pathogenic intronic events and complicated splicing abnormalities in dystrophinopathy
Human Genetics (2023)
-
EMQN best practice guidelines for genetic testing in dystrophinopathies
European Journal of Human Genetics (2020)
-
Exon skipping induced by nonsense/frameshift mutations in DMD gene results in Becker muscular dystrophy
Human Genetics (2020)
-
Mutation Spectrum of Dystrophinopathies in India: Implications for Therapy
The Indian Journal of Pediatrics (2020)