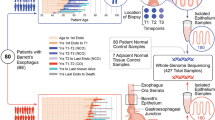
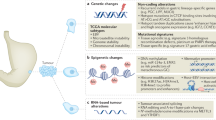
Abstract
Recent studies show that aneuploidy and driver gene mutations precede cancer diagnosis by many years1,2,3,4. We assess whether these genomic signals can be used for early detection and pre-emptive cancer treatment using the neoplastic precursor lesion Barrett’s esophagus as an exemplar5. Shallow whole-genome sequencing of 777 biopsies, sampled from 88 patients in Barrett’s esophagus surveillance over a period of up to 15 years, shows that genomic signals can distinguish progressive from stable disease even 10 years before histopathological transformation. These findings are validated on two independent cohorts of 76 and 248 patients. These methods are low-cost and applicable to standard clinical biopsy samples. Compared with current management guidelines based on histopathology and clinical presentation, genomic classification enables earlier treatment for high-risk patients as well as reduction of unnecessary treatment and monitoring for patients who are unlikely to develop cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data and associated metadata that support the present study have been deposited in the European Genome-phenome Archive under accession number EGAD00001006033. The code and model that support these findings have been provided as an R package in a GitHub repository (https://github.com/gerstung-lab/BarrettsProgressionRisk). Source data are provided with this paper.
References
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020).
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell https://doi.org/10.1016/j.cell.2018.02.020 (2018).
Lee, J. J.-K. et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell https://doi.org/10.1016/j.cell.2019.05.013 (2019).
Abelson, S. et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018).
Gregson, E. M., Bornschein, J. & Fitzgerald, R. C. Genetic progression of Barrett’s oesophagus to oesophageal adenocarcinoma. Br. J. Cancer 115, 403–410 (2016).
Esserman, L. J. et al. Addressing overdiagnosis and overtreatment in cancer: a prescription for change. Lancet Oncol. 15, e234–e242 (2014).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics. CA Cancer J. Clin. 66, 7–30 (2016).
Masclee, G. M. C., Coloma, P. M., De Wilde, M., Kuipers, E. J. & Sturkenboom, M. C. J. M. The incidence of Barrett’s oesophagus and oesophageal adenocarcinoma in the United Kingdom and the Netherlands is levelling off. Aliment. Pharmacol. Ther. 39, 1321–1330 (2014).
Phoa, K. N. et al. Radiofrequency ablation vs endoscopic surveillance for patients with Barrett esophagus and low-grade dysplasia: a randomized clinical trial. J. Am. Med. Assoc. 311, 1209–1217 (2014).
Shaheen, N. J. et al. Radiofrequency ablation in Barrett’s esophagus with dysplasia. N. Engl. J. Med. 360, 2277–2288 (2009).
Parasa, S. et al. Development and validation of a model to determine risk of progression of Barrett’s esophagus to neoplasia. Gastroenterology 154, 1282–1289.e2 (2018).
Younes, M. et al. p53 protein accumulation predicts malignant progression in Barrett’s metaplasia: a prospective study of 275 patients. Histopathology 71, 27–33 (2017).
Pettit, K. & Bellizzi, A. Evaluation of p53 immunohistochemistry staining patterns in Barrett esophagus with low-grade dysplasia. Am. J. Clin. Pathol. 144, A382–A382 (2015).
Sikkema, M. et al. Aneuploidy and overexpression of Ki67 and p53 as markers for neoplastic progression in Barrett’s esophagus: a case–control study. Am. J. Gastroenterol. 104, 2673–2680 (2009).
Keswani, R. N., Noffsinger, A., Waxman, I. & Bissonnette, M. Clinical use of p53 in Barrett’s esophagus. Cancer Epidemiol. Biomark. Prev. 15, 1243–1249 (2006).
Reid, B. J. et al. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am. J. Gastroenterol. 96, 2839–2848 (2001).
Alvi, M. A. et al. DNA methylation as an adjunct to histopathology to detect prevalent, inconspicuous dysplasia and early-stage neoplasia in Barrett’s esophagus. Clin. Cancer Res. 19, 878–888 (2013).
Jin, Z. et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 69, 4112–4115 (2009).
Weaver, J. M. J. et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet. 46, 837–843 (2014).
Secrier, M. et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 48, 1131–1141 (2016).
Frankell, A. M. et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat. Genet. 51, 506–516 (2019).
Nones, K. et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 5, 5224 (2014).
Blum, A. et al. RNA sequencing identifies transcriptionally-viable gene fusions in esophageal adenocarcinomas. Cancer Res. 76, 5587–5589 (2016).
The Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 https://doi.org/10.1038/nature20805 (2017).
Ross-Innes, C. S. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat. Genet. 47, 1038–1046 (2015).
Maley, C. C. et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 38, 468–473 (2006).
Martinez, P. et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat. Commun. 7, 12158 (2016).
Li, X. et al. Assessment of esophageal adenocarcinoma risk using somatic chromosome alterations in longitudinal samples in Barrett’s esophagus. Cancer Prev. Res. 8, 845–856 (2015).
Martinez, P. et al. Evolution of Barrett’s esophagus through space and time at single-crypt and whole-biopsy levels. Nat. Commun. 9, 794 (2018).
Scheinin, I. et al. DNA copy number analysis of fresh and formalin-fixed specimens by whole-genome sequencing: improved correction of systematic biases and exclusion of problematic regions. Genome Res. 24, 2022–2032 (2014).
Li, X. et al. Temporal and spatial evolution of somatic chromosomal alterations: a case–cohort study of Barrett’s esophagus. Cancer Prev. Res. 7, 114–127 (2014).
Shaheen, N. J., Falk, G. W., Iyer, P. G. & Gerson, L. B. ACG clinical guideline: diagnosis and management of Barrett’s esophagus. Am. J. Gastroenterol. 111, 30–50 (2016).
Fitzgerald, R. C. et al. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett’s oesophagus. Gut 63, 7–42 (2014).
Stachler, M. D. et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 47, 1047–1055 (2015).
Kaye, P. V. et al. Novel staining pattern of p53 in Barrett’s dysplasia—the absent pattern. Histopathology 57, 933–935 (2010).
Kaye, P. V. et al. Barrett’s dysplasia and the Vienna classification: reproducibility, prediction of progression and impact of consensus reporting and p53 immunohistochemistry. Histopathology 54, 699–712 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Nilsen, G. et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics 13, 591 (2012).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Acknowledgements
We thank the patients who donated tissue samples to this project. The laboratory of R.C.F. is funded by a Core Programme Grant from the Medical Research Council (grant RG84369). This work was also funded by a United European Gastroenterology Research Prize (RG76026). We thank the Human Research Tissue Bank, which is supported by the UK National Institute for Health Research Cambridge Biomedical Research Centre, from Addenbrooke’s Hospital. Additional infrastructure support was provided from the Cancer Research UK-funded Experimental Cancer Medicine Centre. We also thank B. J. Reid, P. C. Galipeau and C. A. Sanchez from the Fred Hutchinson Cancer Research Center in Seattle, for their time and help in understanding their data, as well as A. Wolfgang Jung from the EMBL-EBI for advice on survival analysis.
Author information
Authors and Affiliations
Contributions
S.K. developed the statistical methods, analyzed data, and wrote the manuscript and supporting information, with input from E.G., R.C.F. and M.G. E.G. gathered the discovery cohort, developed the sWGS methods, generated the sWGS data and curated the clinical information with support from A.V.J. The initial processing pipeline was developed by D.C.W., D.J.W. and M.D.E., and provided input to the data analysis for the sWGS data. W.J., R.d.l.R., C.K. and A.M. identified, collected and assessed pathology for patient samples. S.A., A.B. and C.K. sequenced the validation cohort and quality control samples. R.C.F. initiated and jointly supervised the study with M.G.
Corresponding authors
Ethics declarations
Competing interests
R.C.F. is named on patents for Cytosponge and related assays that have been licensed by the Medical Research Council to Covidien GI Solutions (now Medtronic).
Additional information
Peer review information Javier Carmona was the primary editor on this article, and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Differences in genomic complexity.
a, Per-sample variance in the genomic complexity (cx) values (y-axis) between samples from progressors (n = 424) and non-progressors (n = 349). Boundaries of the box indicate the first and third quartiles of the cx value, horizontal line indicates the median. All data points are shown. While the difference between non-progressors and progressors is significant in a two-sided Wilcoxon rank sum test (p-value=2.4 × 10−6), it only provides limited prognostic signal as the b, ROC curve shows. c, The total number of genomic windows (adjusted by samples per endoscopy) that are CN altered (y-axis) in the 5MB windows and chromosome arms, split by progressor (n = 41) and non-progressor (n = 43) patients at the initial endoscopy. Boundaries of the box indicate the first and third quartiles, of per-patient CN altered counts, center line indicates the median. All data points are shown. Progressors with only a diagnostic endoscopy are excluded. 5MB windows (two-sided Wilcoxon rank sum test, p-value=6 × 10−8) and chromosomal arms (two-sided Wilcoxon rank sum test, p-value 5.54 × 10−11) both show a significant difference in the number of CN alterations identified between the two groups at the initial endoscopy. d, Comparison of chromosome arm altered CN counts (y-axis) found at the initial vs the final endoscopies in progressors and non-progressors. The magnitude of the changes is significantly different between the patient groups (p-value=7 × 10−4, two-sided Wilcoxon rank sum test), demonstrating that alterations to the genomic landscape are apparent in low-resolution WGS data.
Extended Data Fig. 2 Model comparisons for best prediction accuracy.
a, Shows the comparison of the model used in the analysis presented (trained on all samples, n = 773) versus a model which excludes the most extreme histopathological samples (excluding HGD/IMC, n = 711). We compare the accuracy of the ROC AUC using the best sensitivity threshold (Pr = 0.3) presented in Fig. 2a of the main paper. A model trained without use of the extreme samples shows no decrease prediction accuracy indicating that these samples are not driving the differences in the model. b, ROC AUC values describing the prediction accuracy for models trained on different sets of data and various aggregations of per-sample predictions also using the best sensitivity threshold (Pr = 0.3). The first set of bars provides the ROC values for the reference model per-sample predictions (n = 773). The following bars describe the ROC values for aggregated predictions on the same samples: mean and max prediction per endoscopy, mean and max prediction per patient (excluding the final HGD/IMC samples). The aggregated predictions do not differ from the per-sample predictions indicating that a single sample may be sufficient for accurate prediction. All error bars denote the 95% confidence interval for the sensitivity, specificity, and AUC at a threshold of Pr = 0.3.
Extended Data Fig. 3 External validation on Seattle Barrett’s Study SNP data.
Predicting the Seattle Barrett’s Study SNP data using our sWGS CN model results in a lower AUC of 0.77 for all samples (including blood/gastric normals as non-progressor controls) a, Restricted to only BE samples (that is excluding normal), with our higher sensitivity threshold results in an AUC of 0.71 (sensitivity = 0.82, specificity=0.34) b, Overall, the progressor samples show the same pattern of risk classification that the sWGS samples did with high risk classifications occurring at a higher rate in progressive patients independent of pathology. The HGD group in the non-progressor patient group also indicates that our model would classify most of these as progressive. c, Compares ROC values for the SNP data using various additional criteria including: defining patients with HGD as progressed; excluding those with less than 1% of the genome altered (low SCA) and the whole-genome duplicated non-progressor patients (NP WGD); only within the baseline (T1) and penultimate endoscopy (T2) groups respectively. Demonstrating that the model improves as the samples are taken nearer to EAC diagnosis. All error bars denote the 95% confidence interval for the sensitivity, specificity, and AUC at a threshold of Pr = 0.3. d, Plots the mean ratio of the genome altered (y-axis) versus the computationally derived purity value (x-axis) for all timepoint-merged biopsies versus the blood/gastric normal samples. None of the normal samples have more than 1% of the genome altered, and all are >90% purity. Given the issues with assessing very pure, mostly diploid samples, those samples in blue are excluded from the ROC analyses as indicated.
Extended Data Fig. 4 Model trained on only SNP data.
a, Cross-validation classification accuracy at each elastic-net penalty value (penalty = 0 had no non-zero coefficients) for the merged (see Supplementary Information Methods) samples (n = 490) the light blue bar is the penalty value used in the sWGS model and is used for comparison. The numbers on the bars indicate the number of coefficients selected under the given penalty, coefficients in parentheses are those that are stable across 75% of the folds. Error bars show the mean classification accuracy ± s.e.m. a, Volcano plot for the (CVRR) value versus coefficient value for the 27 coefficients from the SNP data trained model. Compared to the coefficients from the sWGS model the (CVRR) values (for example coefficient of variation for the relative risk, see Supplementary Table 3 for definition) are much lower.
Extended Data Fig. 5 Per-patient risk heatmaps.
Samples from the discovery cohort (n = 773) for each progressor patient (n = 45) in a, plotted by the time prior to the final endoscopy (x-axis, endpoint=0) and esophageal location from the sample closest to the esophageal-gastric junction at the bottom up the length of the BE segment, or as many samples as were available for sequencing (y-axis). Each sample is colored by their risk class with shapes inset for each pathology grade. Non-progressor patients are shown in b, (n = 43). These correspond to the mini heatmaps in the main paper Fig. 2.
Extended Data Fig. 6 Increasing numbers of patients improves accuracy.
Analysis showing the potential for improvement by training the model with increasing numbers of patients (x-axis) from the discovery cohort (green and orange bars), combining the discovery and validation cohorts (dark purple bar), and combining all sWGS (discovery and validation) data with the SNP data from the Seattle BE Study (pink bars). In each model we assessed the a, cross-validation accuracy, the b, number of coefficients selected by the model, and finally the c, AUC for a leave-one-out analysis. The green bars are all increasing numbers of patients used in training a model from the discovery cohort (error bars are the mean ± s.e.m. from repeating each training 10 times with randomly selected patients), the orange bar represents the full discovery cohort, the purple bar is the combined discovery and validation (n = 164) cohorts, and the pink bars are the combined sWGS and SNP patients (n = 413).The discovery and validation (all sWGS data) displays consistent improvement in accuracy (0.57 to 0.75) and AUC (0.7 to 0.89) as the number of patients increases. Including the SNP data results in no improvement despite the increased number of patients indicating that the sWGS data alone provides more accurate prognostic information. d, Shows the classification rate per-sample across all 164 patients in the discovery and validation cohorts when we use a model trained on all samples (n = 986). An overall improvement in accuracy for both high and low risk patients is observed.
Extended Data Fig. 7 Cancer risk in relation to p53 IHC per sample.
a, Bars show the proportion of aberrant p53 IHC stained samples separated by pathology in samples from progressive patients. The purple bars indicate the percentage of aberrant samples for each pathology. b, The CN plot from the main paper Fig. 2c zoomed in to chromosome 17 with additional bars shown for the arm-level gains (purple) or losses (green). The blue/yellow outline boxes show the genomic regions that are predictive features of the model. The blue box indicates a loss of 17p arm, while the yellow indicates gain of the 17q arm. Tumor suppressor genes or oncogenes are indicated at their chromosomal location at the bottom of each plot.
Extended Data Fig. 8 Exemplar raw data plots for quality control.
Raw data (red dots) after QDNAseq processing and pcf segmentation (green rectangles), y-axis is the relative GC-adjusted CN value and the x-axis is chromosomal position. The mean absolute deviation (MAD) of the observed (red) versus expected (green) segments was calculated and the variance across the entire sample used to develop a quality cutoff. a, Shows a post-segmentation plot from a cell-line pellet processed into an FFPE block. The wide variance of the raw (red) points results in scattered segmentation (green) high sample mean(MAD) value of 0.015. b, Shows the raw segmented plot from a fresh-frozen EAC tumor. Clear CN alterations can be observed (that is chromosomes 8 and 13). c, Contrasts two different raw data plots from the same FFPE sample in the discovery cohort sequenced as a technical replicate. The sample comes from a non-progressor patient and may have small CN changes that are clearly shared between the two.
Extended Data Fig. 9 Parameter tuning the model.
a, Shows the cross-validation classification accuracy for each bin size (15 kb, 50 kb, 100 kb, 500 kb) at each elastic-net penalty value. Error bars are mean ± s.e.m. of the classification accuracy for each alpha value and bin size. The classification accuracy shows a consistent decline for each bin size. b, Compares the AUC, true positive and false positive rate (TPR, FPR), for each bin size using leave-one-patient-out predictions for the discovery cohort at an elastic-net regression penalty of 0.9. Again, bin size 15 kb shows the best AUC at 0.88, however 50 kb is highly concordant at 0.87. Error bars are the 95% confidence interval. c, Shows the AUC comparison at each bin size for the leave-one-patient-out discovery cohort predictions versus the validation cohort model predictions. At 50 kb the AUCs are 0.87 and 0.84 respectively while all other bin sizes show a much greater difference between the cohorts. Error bars are the 95% confidence interval.
Extended Data Fig. 10 Discretizing risks.
Rate of sample classification by probability discretization per bin size for the a, discovery cohort (n = 773 samples, 88 patients) leave-one-patient-out predictions b, and validation (n = 213 samples, 76 patients) predictions. These confirm that 50 kb is the best parameter to balance classification for type I and type II errors.
Supplementary information
Supplementary Information
Supplementary Note, Methods and Results and Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical source file (b) and all processed copy number data (c–d).
Source Data Fig. 2
Statistical source file.
Source Data Fig. 3
Statistical source file.
Source Data Fig. 4
Statistical source file.
Source Data Extended Data Fig. 1
Statistical source file.
Source Data Extended Data Fig. 2
Statistical source file.
Source Data Extended Data Fig. 3
Statistical source file.
Source Data Extended Data Fig. 4
Statistical source file.
Source Data Extended Data Fig. 5
Statistical source file.
Source Data Extended Data Fig. 6
Statistical source file.
Source Data Extended Data Fig. 7
Statistical source file.
Source Data Extended Data Fig. 8
Labeled QDNA-seq output for each sample in a–c.
Source Data Extended Data Fig. 9
Statistical source file.
Source Data Extended Data Fig. 10
Statistical source file.
Rights and permissions
About this article
Cite this article
Killcoyne, S., Gregson, E., Wedge, D.C. et al. Genomic copy number predicts esophageal cancer years before transformation. Nat Med 26, 1726–1732 (2020). https://doi.org/10.1038/s41591-020-1033-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-1033-y
This article is cited by
-
Multimodal cell-free DNA whole-genome TAPS is sensitive and reveals specific cancer signals
Nature Communications (2025)
-
Context-dependent effects of CDKN2A and other 9p21 gene losses during the evolution of esophageal cancer
Nature Cancer (2025)
-
Transcriptomic changes and gene fusions during the progression from Barrett’s esophagus to esophageal adenocarcinoma
Biomarker Research (2024)
-
Tumor initiation and early tumorigenesis: molecular mechanisms and interventional targets
Signal Transduction and Targeted Therapy (2024)
-
Decoding the basis of histological variation in human cancer
Nature Reviews Cancer (2024)