Abstract
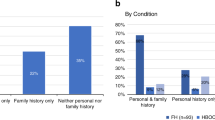
Three inherited autosomal dominant conditions—BRCA-related hereditary breast and ovarian cancer (HBOC), Lynch syndrome (LS) and familial hypercholesterolemia (FH)—have been termed the Centers for Disease Control and Prevention Tier 1 (CDCT1) genetic conditions, for which early identification and intervention have a meaningful potential for clinical actionability and a positive impact on public health1. In typical medical practice, genetic testing for these conditions is based on personal or family history, ethnic background or other demographic characteristics2. In this study of a cohort of 26,906 participants in the Healthy Nevada Project (HNP), we first evaluated whether population screening could efficiently identify carriers of these genetic conditions and, second, we evaluated the impact of genetic risk on health outcomes for these participants. We found a 1.33% combined carrier rate for pathogenic and likely pathogenic (P/LP) genetic variants for HBOC, LS and FH. Of these carriers, 21.9% of participants had clinically relevant disease, among whom 70% had been diagnosed with relevant disease before age 65. Moreover, 90% of the risk carriers had not been previously identified, and less than 19.8% of these had documentation in their medical records of inherited genetic disease risk, including family history. In a direct follow-up survey with all carriers, only 25.2% of individuals reported a family history of relevant disease. Our experience with the HNP suggests that genetic screening in patients could identify at-risk carriers, who would not be otherwise identified in routine care.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors.
Code availability
The code that supports the findings of this study is available from the corresponding authors upon reasonable request.
References
Centers for Disease Control and Prevention, Office of Public Health Genomics. Tier 1 genomic applications toolkit for public health departments. https://www.cdc.gov/genomics/implementation/toolkit/index.htm (2019).
Khoury, M. J. et al. A collaborative translational research framework for evaluating and implementing the appropriate use of human genome sequencing to improve health. PLoS Med. 15, e1002631 (2018).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423 (2015).
Dewey, F. E. et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354, aaf6814 (2016).
Abul-Husn, N. S. et al. Genetic identification of familial hypercholesterolemia within a single US health care system. Science 354, aaf7000 (2016).
Manickam, K. et al. Exome sequencing–based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw. Open 1, e182140 (2018).
National Comprehensive Cancer Network. Genetic/ Familial High-Risk Assessment: Breast and Ovarian. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) https://www2.tri-kobe.org/nccn/guideline/gynecological/english/genetic_familial.pdf (2019).
National Comprehensive Cancer Network. Genetic/ Familial High-Risk Assessment: Colorectal. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) https://www2.tri-kobe.org/nccn/guideline/colorectal/english/genetics_colon.pdf (2017).
Sturm, A. C. et al. Clinical genetic testing for familial hypercholesterolemia. J. Am. Coll. Cardiol. 72, 662–680 (2018).
Grundy, S. M. et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. J. Am. Coll. Cardiol. 73, e285–e350 (2019).
Versmissen, J. et al. Efficacy of statins in familial hypercholesterolaemia: a long-term cohort study. BMJ 337, a2423 (2008).
Kohlmann, W. & Gruber, S. B. Lynch syndrome. In GeneReviews (eds. Adam, M. P. et al.) (University of Washington, 2018).
Senter, L. et al. The clinical phenotype of Lynch syndrome due to germline PMS2 mutations. Gastroenterology 135, 419–428 (2008).
Fahed, A. C. & Nemer, G. M. Familial hypercholesterolemia: the lipids or the genes? Nutr. Metab. 8, 23 (2011).
Chen, S. & Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 25, 1329–1333 (2007).
Kuchenbaecker, K. B. et al. Risks of breast, ovarian and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017).
National Academies of Sciences, Engineering and Medicine. Communities in Action: Pathways to Health Equity (eds. Baciu, A. et al.) (National Academies Press, 2017).
Iacocca, M. A. et al. Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia. J. Lipid Res. 58, 2202–2209 (2017).
Schmidt, A. Y. et al. Next-generation sequencing-based detection of germline copy number variations in BRCA1/BRCA2: validation of a one-step diagnostic workflow. J. Mol. Diagn. 19, 809–816 (2017).
Albert L Siu, A.L. & U.S. Preventive Services Task Force. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 164, 279–296 (2016).
US Preventive Services Task Force et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. J. Am. Med. Assn. 315, 2564–2575 (2016).
Bennette, C. S., Gallego, C. J., Burke, W., Jarvik, G. P. & Veenstra, D. L. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet. Med. 17, 587–595 (2015).
Zhang, L. et al. Population genomic screening of all young adults in a health-care system: a cost-effectiveness analysis. Genet. Med. 21, 1958–1968 (2019).
Andermann, A., Blancquaert, I., Beauchamp, S. & Déry, V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull. World Health Organ. 86, 317–319 (2008).
Grzymski, J. J. et al. The Healthy Nevada Project: rapid recruitment for population health study. Preprint at bioRxiv https://doi.org/10.1101/250274 (2018).
Helix. Personal Genomics Platform White Paper. https://cdn.shopify.com/s/files/1/0004/5416/4542/files/Helix-Performance-White-Paper_v2.pdf?3324109126506576509 (2017).
Biesecker, L. G. & Harrison, S. M., ClinGen Sequence Variant Interpretation Working Group. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet. Med. 20, 1687–1688 (2018).
Tyrer, J., Duffy, S. W. & Cuzick, J. A breast cancer prediction model incorporating familial and personal risk factors. Stat. Med. 23, 1111–1130 (2004).
Lindor, N. M. et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of Penn II model to previous study. Fam. Cancer 9, 495–502 (2010).
Haase, A. & Goldberg, A. C. Identification of people with heterozygous familial hypercholesterolemia. Curr. Opin. Lipidol. 23, 282–289 (2012).
Acknowledgements
J.J.G., G.E., E.S., K.A.S., R.R., W.J.M., B.L. and H.R. were supported by grants from the Nevada Governor’s Office of Economic Development Knowledge Fund (no. 14685) and by Renown Health (no. GR09183). We thank all participants in the HNP. We also thank the operational teams of the HNP and Helix.
Author information
Authors and Affiliations
Contributions
J.J.G., G.E., A.S., A.B. and J.T.L. conceived the presented idea and contributed to the analysis and manuscript writing. J.A.M.R. and M.F. performed P/LP variant interpretation for all participants. C.R., N.S., E.L. and L.S. contributed clinical oversight and clinical data gathering. E.S., K.A.S., J.K., W.J.M., S.D., H.R., B.L., M.R., J.B., K.D. and N.W. helped to organize both clinical and genomic data and execution of the study. J.J.G. and J.T.L. oversaw the overall program. All authors provided critical feedback and helped to shape the research, analysis and manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.S., E.L., J.K., M.R., J.B., K.D., N.W., A.B. and J.T.L. are employees of Helix. J.J.G., G.E., J.A.M.R., K.A.S., R.R., C.R., N.S., S.D., W.J.M., B.L., H.R., A.S. and M.F. declare no competing interests.
Additional information
Peer review information Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Carrying a P/LP variant in a FH gene leads to increased risk and earlier onset of hyperlipidemia and treatment with anti-hyperlipidemic medications.
Disease-free survival for ‘carriers’ (n = 73) of a FH P/LP variant and ‘non-FH carriers’ (n = 20,117) for a, first report of a ICD-10 code for hyperlipidemia in the medical record, and b, first order for an anti-hyperlipidemic medication in the medical record. Solid colored lines (and the matching shaded regions) are survival curves with 95% confidence intervals. The dotted lines represent the median survival age. p-values represent the results of a 2-sided log-rank test. No adjustments were made for multiple comparisons.
Extended Data Fig. 2 Empirical cumulative distribution for age of first diagnosis for P/LP carriers with relevant disease diagnoses.
Clinical relevant disease manifestation for at-risk carriers in our cohort occurs between the ages of 24 and 80, with approximately 40% of disease occurring before the age of 50.
Extended Data Fig. 3 Comparison of the clinical impact of P/LP variants in different genes for the same CDC Tier1 genomic condition.
a-d, Disease-free survival (DFS) for carriers (n = 273) in the Healthy Nevada Project for individuals with medical records at Renown Health. Data are shown for P/LP variant carriers for BRCA1 (n = 54) and BRCA2 (n = 81) for HBOC (a), for P/LP variant carriers for MSH6 (n = 23) and PMS2 (n = 32) for all-cause cancers. There was not sufficient data to explore MLH1 or MSH2 independently (b), for P/LP variant carriers for APOB (n = 14) and LDLR (n = 58) for presentation of hyperlipidemia and (d) for treatment with anti-hyperlipidemic medication. There was not sufficient data to explore PCSK9 independently. Solid colored lines (and the matching shaded regions) are survival curves with 95% confidence intervals. The dotted lines represent the median survival age. p-values represent the results of a 2-sided log-rank test. No adjustments were made for multiple comparisons.
Extended Data Fig. 4 Individuals who likely fall within existing FH clinical guidelines do not have risk in excess of carrying a P/LP variant.
Disease-free survival (DFS) for carriers (n = 273) that likely fall ‘within guidelines’ and those ‘outside guidelines.’ Data are shown for within guidelines (n = 10) vs outside guidelines (n = 63) for FH P/LP variant carriers for presentation of hyperlipidemia a, and for treatment with anti-hyperlipidemic agents b. Solid colored lines (and the matching shaded regions) are survival curves with 95% confidence intervals. The dotted lines represent the median survival age. p-values represent the results of a 2-sided log-rank test. No adjustments were made for multiple comparisons.
Supplementary information
Rights and permissions
About this article
Cite this article
Grzymski, J.J., Elhanan, G., Morales Rosado, J.A. et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med 26, 1235–1239 (2020). https://doi.org/10.1038/s41591-020-0982-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-0982-5