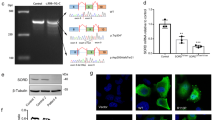
Abstract
Here we report biallelic mutations in the sorbitol dehydrogenase gene (SORD) as the most frequent recessive form of hereditary neuropathy. We identified 45 individuals from 38 families across multiple ancestries carrying the nonsense c.757delG (p.Ala253GlnfsTer27) variant in SORD, in either a homozygous or compound heterozygous state. SORD is an enzyme that converts sorbitol into fructose in the two-step polyol pathway previously implicated in diabetic neuropathy. In patient-derived fibroblasts, we found a complete loss of SORD protein and increased intracellular sorbitol. Furthermore, the serum fasting sorbitol levels in patients were dramatically increased. In Drosophila, loss of SORD orthologs caused synaptic degeneration and progressive motor impairment. Reducing the polyol influx by treatment with aldose reductase inhibitors normalized intracellular sorbitol levels in patient-derived fibroblasts and in Drosophila, and also dramatically ameliorated motor and eye phenotypes. Together, these findings establish a novel and potentially treatable cause of neuropathy and may contribute to a better understanding of the pathophysiology of diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data described in this paper are present either in the main text or in the Supplementary Information. Source data for Fig. 2 are presented with the paper. The sequence data obtained by WES and WGS are not publicly available because the study participants did not give full consent for releasing data publicly.
Change history
26 May 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41588-020-0649-7
References
Rossor, A. M., Tomaselli, P. J. & Reilly, M. M. Recent advances in the genetic neuropathies. Curr. Opin. Neurol. 29, 537–548 (2016).
Fridman, V. et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J. Neurol. Neurosurg. Psychiatry 86, 873–878 (2015).
Cortese, A. et al. Targeted next-generation sequencing panels in the diagnosis of Charcot-Marie-Tooth disease. Neurology 94, e51–e61 (2020).
Gonzalez, M. et al. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum. Mutat. 36, 950–956 (2015).
Hellgren, M., Kaiser, C., de Haij, S., Norberg, A. & Höög, J.-O. A hydrogen-bonding network in mammalian sorbitol dehydrogenase stabilizes the tetrameric state and is essential for the catalytic power. Cell. Mol. Life Sci. 64, 3129–3138 (2007).
Carr, A. S. et al. A study of the neuropathy associated with transthyretin amyloidosis (ATTR) in the UK. J. Neurol. Neurosurg. Psychiatry 87, 620–627 (2016).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Lazarin, G. A. et al. An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet. Med. 15, 178–186 (2013).
Antonarakis, S. E. Carrier screening for recessive disorders. Nat. Rev. Genet. 20, 549–561 (2019).
Murphy, S. M. et al. Reliability of the CMT neuropathy score (second version) in Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst. 16, 191–198 (2011).
Johansson, K. et al. Crystal structure of sorbitol dehydrogenase. Chem. Biol. Interact. 130–132, 351–358 (2001).
Lindstad, R. I., Teigen, K. & Skjeldal, L. Inhibition of sorbitol dehydrogenase by nucleosides and nucleotides. Biochem. Biophys. Res. Commun. 435, 202–208 (2013).
Luque, T. et al. Sorbitol dehydrogenase of Drosophila. Gene, protein, and expression data show a two-gene system. J. Biol. Chem. 273, 34293–34301 (1998).
Bellen, H. J. et al. The Drosophila Gene Disruption Project: progress using transposons with distinctive site specificities. Genetics 188, 731–743 (2011).
Bausenwein, B., Dittrich, A. P. & Fischbach, K. F. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 267, 17–28 (1992).
Kikkawa, R. et al. Effect of a new aldose reductase inhibitor, (E)-3-carboxymethyl-5-[(2E)-methyl-3-phenylpropenylidene]rhodanine (ONO-2235) on peripheral nerve disorders in streptozotocin-diabetic rats. Diabetologia 24, 290–292 (1983).
Matsumoto, T. et al. Long-term treatment with ranirestat (AS-3201), a potent aldose reductase inhibitor, suppresses diabetic neuropathy and cataract formation in rats. J. Pharmacol. Sci. 107, 340–348 (2008).
Ramirez, M. A. & Borja, N. L. Epalrestat: an aldose reductase inhibitor for the treatment of diabetic neuropathy. Pharmacotherapy 28, 646–655 (2008).
Hao, W. et al. Hyperglycemia promotes Schwann cell de-differentiation and de-myelination via sorbitol accumulation and Igf1 protein down-regulation. J. Biol. Chem. 290, 17106–17115 (2015).
Grewal, A. S., Bhardwaj, S., Pandita, D., Lather, V. & Sekhon, B. S. Updates on aldose reductase inhibitors for management of diabetic complications and non-diabetic diseases. Mini Rev. Med. Chem. 16, 120–162 (2016).
Chalk, C., Benstead, T. J. & Moore, F. Aldose reductase inhibitors for the treatment of diabetic polyneuropathy. Cochrane Database Syst. Rev. 17, CD004572 (2007).
Polydefkis, M. et al. Safety and efficacy of ranirestat in patients with mild-to-moderate diabetic sensorimotor polyneuropathy. J. Peripher. Nerv. Syst. 20, 363–371 (2015).
Sekiguchi, K. et al. Aldose reductase inhibitor ranirestat significantly improves nerve conduction velocity in diabetic polyneuropathy: a randomized double-blind placebo-controlled study in Japan. J. Diabetes Investig. 10, 466–474 (2019).
Züchner, S. et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36, 449–451 (2004).
De Vos, M., Hayward, B. E., Picton, S., Sheridan, E. & Bonthron, D. T. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am. J. Hum. Genet. 74, 954–964 (2004).
Rumsby, G., Carroll, M. C., Porter, R. R., Grant, D. B. & Hjelm, M. Deletion of the steroid 21-hydroxylase and complement C4 genes in congenital adrenal hyperplasia. J. Med. Genet. 23, 204–209 (1986).
Chen, J.-M., Cooper, D. N., Chuzhanova, N., Férec, C. & Patrinos, G. P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775 (2007).
Harel, T. et al. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 99, 831–845 (2016).
Lupski, J. R. et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66, 219–232 (1991).
Schmidt, R. E. et al. Inhibition of sorbitol dehydrogenase exacerbates autonomic neuropathy in rats with streptozotocin-induced diabetes. J. Neuropathol. Exp. Neurol. 60, 1153–1169 (2001).
Schmidt, R. E. et al. A potent sorbitol dehydrogenase inhibitor exacerbates sympathetic autonomic neuropathy in rats with streptozotocin-induced diabetes. Exp. Neurol. 192, 407–419 (2005).
Obrosova, I. G. Increased sorbitol pathway activity generates oxidative stress in tissue sites for diabetic complications. Antioxid. Redox Signal. 7, 1543–1552 (2005).
Sango, K. et al. High glucose-induced activation of the polyol pathway and changes of gene expression profiles in immortalized adult mouse Schwann cells IMS32. J. Neurochem. 98, 446–458 (2006).
Holmes, R. S., Duley, J. A. & Hilgers, J. Sorbitol dehydrogenase genetics in the mouse: a “null” mutant in a “European” C57BL strain. Anim. Blood Groups Biochem. Genet. 13, 263–272 (1982).
Lee, A. Y., Chung, S. K. & Chung, S. S. Demonstration that polyol accumulation is responsible for diabetic cataract by the use of transgenic mice expressing the aldose reductase gene in the lens. Proc. Natl Acad. Sci. USA 92, 2780–2784 (1995).
Ng, T. F. et al. Effects of sorbitol dehydrogenase deficiency on nerve conduction in experimental diabetic mice. Diabetes 47, 961–966 (1998).
Ruff, J. S. et al. Human-relevant levels of added sugar consumption increase female mortality and lower male fitness in mice. Nat. Commun. 4, 2245 (2013).
Callaghan, B. C., Cheng, H. T., Stables, C. L., Smith, A. L. & Feldman, E. L. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 11, 521–534 (2012).
Dyck, P. J. et al. The prevalence by staged severity of various types of diabetic neuropathy, retinopathy, and nephropathy in a population-based cohort: the Rochester Diabetic Neuropathy Study. Neurology 43, 817–824 (1993).
Lorenzi, M. The polyol pathway as a mechanism for diabetic retinopathy: attractive, elusive, and resilient. Exp. Diabetes Res. 2007, 61038 (2007).
Li, L. et al. The induction of trehalose and glycerol in Saccharomyces cerevisiae in response to various stresses. Biochem. Biophys. Res. Commun. 387, 778–783 (2009).
Acknowledgements
This project was supported by the NINDS (R01NS075764 to S.Z. and M.S.; R01NS105755 to S.Z.), the NIH (R21GM119018 and 1R61AT010408 to R.G.Z.), the NCATS (U54NS065712 to M.S.), the CMT Association, the Hereditary Neuropathy Foundation, The Genesis Project foundation, the Muscular Dystrophy Association, the European Union’s Horizon 2020 research and innovation programme under the ERA-NET Cofund action no. 643578 under the frame of the E-Rare-3 network PREPARE (01GM1607 to M.S.; and unfunded to S.Z.), the grant 779257 ‘Solve-RD’ (to R.S. and M.S., M.M.R. and H.H.) and the National Institute for Health Research University College London Hospitals Biomedical Research Centre (to M.L.). The project received further support from the ‘Bundesministerium für Bildung und Forschung’ (BMBF) via funding for the TreatHSP consortium (01GM1905 to R.S.) and the National Institutes of Health (grant 5R01NS072248 to R.S. and S.Z.), the Austrian Science Fund (FWF, P27634FW to M.A.-G.) and the National Natural Science Foundation of China (81771366). A.C. thanks the Medical Research Council (MR/T001712/1), the Wellcome Trust (204841/Z/16/Z), the Fondazione CARIPLO (2019-1836), the Italian Ministry of Health Ricerca Corrente 2018–2019 and the Inherited Neuropathy Consortium (INC) for grant support. H.H. and M.M.R. thank the MRC, the Wellcome Trust, the MDA, MD UK, Ataxia UK, The MSA Trust, the Rosetrees Trust and the NIHR UCLH BRC for grant support. A.M.R. is funded by a Wellcome Trust Postdoctoral Fellowship for Clinicians (110043/Z/15/Z). D.N.H. receives grant support through NIH U54 NS065712-09, the Muscular Dystrophy Association, the Friedreich’s Ataxia Alliance and Voyager Pharmaceuticals. We thank M. Tekin for kindly providing DNA from healthy controls or Turkish ancestry. We also thank Twenty Three Calvin (Marie Stargala and Matthew Rosen) for creating the cover art for the issue.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization: A.C. and S.Z. Funding acquisition: S.Z., M.E.S., M.M.R., H.H., R.S. and M.S., Investigation: A.C., Y.Z., A.P.R., S.N., S.C., M.P., E.Buglo, R.G.Z. and S.Z. Resources: A.C., Y.Z., A.P.R., S.N., S.C., L.A., A.A.-A, M.A.-G., C.J.B., Y.B., D.M.B-.B., E.Bugiardini, J.D., M.C.D., S.M.E.F., A.A.-F., E.G., M.A.A., S.A.H., N.A.H., H.H., R.I., A.K., M.L., Z.L., S.M., T.M., F.M., E.M., D.P., M.P., C.P., E.P., A.M.R., L.S., S.S.S., R.S., J.E.S., T.S., M.S., P.S., B.T., F.T., S.T., J.V., R.Z., D.N.H., M.M.R., M.E.S., R.G.Z. and S.Z. Supervision: S.Z. and R.G.Z. Writing–original draft: A.C., Y.Z., A.P.R., R.G.Z. and S.Z. All authors contributed to revising the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
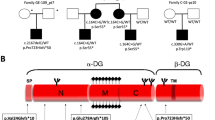
Extended Data Fig. 1 Pedigrees of families carrying biallelic mutations in SORD.
The squares indicate males, the circles females, and the diagonal lines deceased individuals. Patients are indicated with filled shapes. Genotypes are provided when tested by Sanger sequencing.
Extended Data Fig. 2 Loss of Drosophila Sodh does not affect life span.
Life span of control flies (yw) and Sodh2MB01265/MB01265 flies. Data are shown in Kaplan-Meier survival plot. n = 100 biologically independent animals. Significance level was established by a two-sided log-rank test.
Extended Data Fig. 3 Double knockdown of Drosophila Sodh1 and Sodh2 causes age-dependent synaptic degeneration.
a,b, Laminae of control (GMR-GAL4 heterozygotes) or Sodh1 and Sodh2 double knockdown homozygous flies at 2 DAE and 10 DAE were stained with HRP (green; marks neuronal membranes) and BRP (magenta; marks synaptic active zones). Yellow arrowheads indicate vacuole-like structures in the lamina that correspond to missing terminals. The areas outlined by yellow boxes are shown at higher magnification. The intensity of BRP is indicated using a red spectrum. Dotted lines indicate the area of lamina vacuole-like structures. Scale bars: 30 μm. c, Quantification of the number and size of vacuole-like structures. n = 8 biologically independent samples. Data are presented as mean ± s.d. (error bars). Statistical analysis was performed using two-way ANOVA followed by post-hoc Tukey’s multiple comparison test.
Extended Data Fig. 4 Treatment with aldose reductase inhibitors Epalrestat and Ranirestat restore locomotor function in Sodh1 and Sodh2 double knockdown flies.
Locomotor activity of control flies (yw) feeding with DMSO, or flies with neuronal-specific knockdown of Sodh1 and Sodh2 feeding with DMSO, 80 μg/ml Epalrestat, or 80 μg/ml Ranirestat. n = 10 in 2, 10, 20, 30, 40 DAE, and n = 8 in 50 DAE. Data are presented as mean ± s.d. (error bars). Statistical analysis was performed using two-way ANOVA followed by post-hoc Tukey’s multiple comparison test.
Supplementary information
Supplemental Information
Supplementary Tables 1–4
Source data
Source Data Fig. 2
Unprocessed western blots for Fig. 2.
Rights and permissions
About this article
Cite this article
Cortese, A., Zhu, Y., Rebelo, A.P. et al. Biallelic mutations in SORD cause a common and potentially treatable hereditary neuropathy with implications for diabetes. Nat Genet 52, 473–481 (2020). https://doi.org/10.1038/s41588-020-0615-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0615-4