Abstract
Inflammatory bowel diseases involve the dynamic interaction of host genetics, the microbiome and inflammatory responses. Here we found lower expression of NLRP12 (which encodes a negative regulator of innate immunity) in human ulcerative colitis, by comparing monozygotic twins and other patient cohorts. In parallel, Nlrp12 deficiency in mice caused increased basal colonic inflammation, which led to a less-diverse microbiome and loss of protective gut commensal strains (of the family Lachnospiraceae) and a greater abundance of colitogenic strains (of the family Erysipelotrichaceae). Dysbiosis and susceptibility to colitis associated with Nlrp12 deficency were reversed equally by treatment with antibodies targeting inflammatory cytokines and by the administration of beneficial commensal Lachnospiraceae isolates. Fecal transplants from mice reared in specific-pathogen-free conditions into germ-free Nlrp12-deficient mice showed that NLRP12 and the microbiome each contributed to immunological signaling that culminated in colon inflammation. These findings reveal a feed-forward loop in which NLRP12 promotes specific commensals that can reverse gut inflammation, while cytokine blockade during NLRP12 deficiency can reverse dysbiosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Referenced accessions
European Nucleotide Archive
Gene Expression Omnibus
Change history
17 April 2017
In the version of this article initially published, the second sentence in the first subsection of Results incorrectly described an experimental group as "...seven additional patients with UC...". The correct description is "...seven additional UC patient cohorts...". The error has been corrected in the HTML and PDF versions of the article.
18 May 2017
In the version of this article initially published, the vertical axis of Figure 5j was incorrectly labeled 'PC1 (22.56%)'. The correct label is 'PC3 (6.47%)'. The error has been corrected in the HTML and PDF versions of this article.
18 October 2017
Nat. Immunol. 18, 541–551 (2017); published 13 March 2017; corrected after print 17 April 2017; corrected after print 18 May 2017 In the version of this article initially published, the vertical axis of Figure 5j was incorrectly labeled 'PC1 (22.56%)'. The correct label is 'PC3 (6.47%)'. The error has been corrected in the HTML and PDF versions of this article.
References
Kau, A.L., Ahern, P.P., Griffin, N.W., Goodman, A.L. & Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Karrasch, T. & Jobin, C. NF-κB and the intestine: friend or foe? Inflamm. Bowel Dis. 14, 114–124 (2008).
Hugot, J.P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013).
Ramanan, D., Tang, M.S., Bowcutt, R., Loke, P. & Cadwell, K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity 41, 311–324 (2014).
Jiang, W. et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med. 210, 2465–2476 (2013).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Vladimer, G.I. et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96–107 (2012).
Williams, K.L. et al. The CATERPILLER protein monarch-1 is an antagonist of Toll-like receptor-, tumor necrosis factor α-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 280, 39914–39924 (2005).
Lich, J.D. et al. Monarch-1 suppresses non-canonical NF-κB activation and p52-dependent chemokine expression in monocytes. J. Immunol. 178, 1256–1260 (2007).
Allen, I.C. et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity 36, 742–754 (2012).
Zaki, M.H. et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell 20, 649–660 (2011).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014).
Sartor, R.B. & Wu, G.D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 152, 327–339 (2016).
Frank, D.N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104, 13780–13785 (2007).
Kitajima, S., Morimoto, M., Sagara, E., Shimizu, C. & Ikeda, Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp. Anim. 50, 387–395 (2001).
Zaki, M.H., Man, S.M., Vogel, P., Lamkanfi, M. & Kanneganti, T.D. Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proc. Natl. Acad. Sci. USA 111, 385–390 (2014).
Gallo, R.L. & Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 (2012).
Kuczynski, J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. in Current Opinions in Microbiology Chapter 1, Unit 1E.5. (John Wiley & Sons, 2012).
Carmody, R.N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Baumgart, M. et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1, 403–418 (2007).
Ridaura, V.K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Reeves, A.E., Koenigsknecht, M.J., Bergin, I.L. & Young, V.B. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun. 80, 3786–3794 (2012).
Denning, T.L., Wang, Y.C., Patel, S.R., Williams, I.R. & Pulendran, B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8, 1086–1094 (2007).
Cerovic, V., Bain, C.C., Mowat, A.M. & Milling, S.W. Intestinal macrophages and dendritic cells: what's the difference? Trends Immunol. 35, 270–277 (2014).
Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Peyrin-Biroulet, L. Anti-TNF therapy in inflammatory bowel diseases: a huge review. Minerva Gastroenterol. Dietol. 56, 233–243 (2010).
Jones, S.A., Scheller, J. & Rose-John, S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 121, 3375–3383 (2011).
Tanaka, T., Narazaki, M. & Kishimoto, T. Anti-interleukin-6 receptor antibody, tocilizumab, for the treatment of autoimmune diseases. FEBS Lett. 585, 3699–3709 (2011).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Kawamoto, S. et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014).
Sonnenburg, E.D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633 (2014).
Schubert, A.M. et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio 5, e01021–e01146 (2014).
Nava, G.M., Friedrichsen, H.J. & Stappenbeck, T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011).
Reichardt, N. et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335 (2014).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Smith, P.M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Handley, S.A. et al. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19, 323–335 (2016).
Dinh, D.M. et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211, 19–27 (2015).
Schmidt, R. et al. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5, 64 (2014).
Geem, D., Medina-Contreras, O., Kim, W., Huang, C.S. & Denning, T.L. Isolation and characterization of dendritic cells and macrophages from the mouse intestine. J. Vis. Exp. 63, 4040 (2012).
Seo, S.U. et al. Distinct Commensals Induce Interleukin-1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity 42, 744–755 (2015).
Stevenson, B.S., Eichorst, S.A., Wertz, J.T., Schmidt, T.M. & Breznak, J.A. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70, 4748–4755 (2004).
Berry, D. et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 6, 2091–2106 (2012).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Acknowledgements
We thank the following for core and technical support: the University of North Carolina (UNC)–North Carolina State University Center for Gastrointestinal Biology and Disease; the UNC Microbiome Core Facility; S.L. Tonkonogy and the Gnotobiotic Core at North Carolina State University; M.A. Bower and J.W. Herzog at the National Gnotobiotic Rodent Resource Center and Crohn's and Colitis Foundation of America Gnotobiotic Animal Facility at UNC; the UNC Lineberger Comprehensive Cancer Center Animal Histopathology Core Facility and Animal Studies Core Facility; and the UNC Flow Cytometry Core Facility. Supported by the US National Institute of Health (RO1-CA156330 to J.P.-Y.T. (National Cancer Institute); P01 DK094779 to J.P.-Y.T., P30 DK034987 to R.B.S., F32-DK088417-01 to J.E.W. and F32-DK098916 to A.D.T. (National Institute of Diabetes and Digestive and Kidney Diseases); U19-AI067798 and R37 AI029564 to J.P.-Y.T. and U19 AI090871 to V.B.Y. (National Institute of Allergy and Infectious Disease); and P40 OD010995 to R.B.S. (Office of the Director)), the American Cancer Society (PF-13-401-01-TBE to J.E.W.), the Crohn's and Colitis Foundation of America (R.B.S.) and the National Multiple Sclerosis Society (FG 1968-A-1 to W-C.C.).
Author information
Authors and Affiliations
Contributions
L.C., J.E.W. and J.P.-Y.T. designed the experiments and wrote the manuscript, with critical input from V.B.Y. and R.B.S.; M.J.K. and V.B.Y. generated the purified Lachnospiraceae strains; W.-C.C. contributed to the immunoblot analysis, cytokine measurement and flow cytometry; S.A.M. performed the histopathological scoring; A.D.T., W.J.B. and G.K.M. generated the radiation-bone-marrow chimeras; C.D.P., N.M., S.E.P. and R.B.S. contributed to the isolation of fecal DNA and 16S rRNA gene-sequencing experiments; and R.B.S. generated the GF mice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
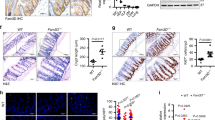
Supplementary Figure 1 NLRP12 is downregulated in biopsy samples from patients with active ulcerative colitis (UC).
(a-g) NLRP12 expression in 7 UC mRNA profiling studies deposited in NCBI GEO database. Disease stages, GEO accession numbers, subject tissues for mRNA purification, therapeutic treatments including corticosteroid or anti-TNF (infliximab) and mRNA measuring microarrays and sequencer platforms are listed. For (a), n=5/group; (b), healthy (n=6), UC nonresponse (n=13), UC response (n=8); (c) healthy (n=20), active UC (n=15), inactive UC (n=18); (d) healthy (n=12), active UC (n=13); (e) n=20/group; (f) healthy (n=6), UC before treatment (n=20), UC response to treatment (n=8); (g) healthy (n=42), UC (n=40). Error bars show SEM. *p<0.05, **p<0.01 and n.s. means no significance determined by two-tailed unpaired t test.
Supplementary Figure 2 Conventionally raised Nlrp12–/– mice display exacerbated colitis.
(a) DAI and (b) colon length (n=17/group) of conventionally-raised mice treated with 3% DSS, compiled from 3 independent experiments. (c) Representative images of H&E-stained colons from conventionally-raised or germ-free mice after DSS-induced colitis. Scale bars represent 1 mm for 40X and 200 μm for 200X. (d) Histopathology scoring of H&E-stained colons (WT, n=7; Nlrp12–/–, n=8; GF WT, n=6; GF Nlrp12–/–, n=6). (e-g) Representative immunoblots of distal colon proteins and composite densitometry from 2 independent experiments. One dot or one lane represents one mouse. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, and n.s. means no significance determined by two-tailed unpaired t test.
Supplementary Figure 3 Nlrp12 defieciency causes significant changes in the intestinal microbiota.
(a) Schematic showing fecal DNA collection from mice housed in two distinct vivarium for 16s rRNA gene sequencing. (b) Bacterial diversity within WT (n=18) and Nlrp12–/– (n=15) mice, (c) PCoA plot showing microbiota compositional differences between WT and Nlrp12-/- mice from a repeated 16S rRNA gene microbiome sequencing experiment conducted in vivarium #2. (d) Schematic showing fecal DNA collection from Nlrp12+/+ and Nlrp12–/– littermates derived from the same parents for 16s rRNA gene sequencing. (e) PCoA plots of microbiota compositional differences between Nlrp12+/+ and Nlrp12–/–littermates. Error bars show SEM. One dot represents one mouse. **p<0.01 determined by two-tailed unpaired t test.
Supplementary Figure 4 Nlrp12–/– mice cohoused with wild-type mice display attenuated colitis.
(a) Representative images of H&E-stained colons from DSS-treated cohoused or single-housed WT and Nlrp12–/– mice. (b) Body weight, (c) percent survival († indicates statistical significance between SiHo WT vs. SiHo Asc–/–, and * indicates significance between SiHo WT vs. CoHo WT), (d) DAI and (e) colon length of DSS-treated cohoused or single-housed WT and Asc–/– mice (CoHo WT and CoHo Asc–/–, n=4; SiHo WT and SiHo Asc–/–, n=6). (f) Representative images of H&E-stained colons of SiHo and Coho WT and Asc–/– mice after DSS-induced colitis. (g) PCoA plot showing fecal microbial composition before cohousing (n=9/group). (h) Quantification of UniFrac distance between mice from (g) after cohousing, indicated as dissimilarity values. Error bars show SEM. *p<0.05, ***p<0.001 and n.s. means no significance, determined by two-tailed unpaired t test (b, d, e), Log-rank (Mantel Cox) test (c) and ANOSIM (h). Scale bars represent 1 mm for 40X and 200 μm for 200X (a,f). The Tukey’s boxplot indicates first (bottom of the box) and third quartiles (top of the box), and the band inside the box is the median, and the ends of the whiskers indicate 1.5 interquartile range of the upper or lower quartile (h).
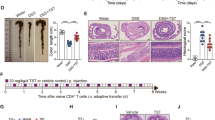
Supplementary Figure 5 Inoculation of Lachnospiraceae strains suppresses colitis in Nlrp12–/– mice.
(a) Representative images of H&E-stained colons from mice administered Lachnospiraceae or BHI vehicle prior to DSS treatment. Scale bars represent 1 mm for 40X and 200 μm for 200X.
Supplementary Figure 6 Treatment with anti-TNF and anti-IL6R reverses dysbiosis in Nlrp12–/– mice.
(a, b) Flow chart showing WT and Nlrp12–/– littermates treated with anti-TNF and anti-IL6R antibodies (Ab) or PBS, and experimental strategy for Ab dosing (filled triangles) and fecal collection (open triangles) (n=6 mice/group). (c) Microbial diversity and (d) PCoA plots of microbial compositional differences in WT and Nlrp12–/– mice before antibody treatment. (e) Quantification of UniFrac distances calculated from (d) represented as dissimilarity values. Error bars show SEM. *p<0.05, **p<0.01, ****p<0.0001 and n.s. means no significance determined by two-tailed unpaired t test (c) or ANOSIM (e).
Supplementary Figure 7 Model for the role of NLRP12 in maintaining intestinal homeostasis between host innate immunity and intestinal commensal symbiosis.
Graphic summary of NLRP12 in maintaining intestinal homeostasis between host innate immunity and intestinal commensal symbiosis. The dysbiosis and inflammation caused by dysfunction of NLRP12 are shown in red. And therapeutic intervention points for the treatment of IBD during NLRP12 dysfunction are shown in blue.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1909 kb)
Supplementary Table 1
Microbiome sequencing of fecal DNA from Wild Type and Nlrp12–/– mice housed in vivarium #1 (XLS 135 kb)
Supplementary Table 2
Microbiome sequencing of fecal DNA from Wild Type and Nlrp12–/– mice housed in vivarium #2. (XLSX 188 kb)
Supplementary Table 4
Nlrp12–/– mice and IBD patients share similar reduction of specific microbiota. (XLSX 1061 kb)
Supplementary Table 6
The specificity and coverage of the primers used to identify the 23 Lachnospiraceae strains (XLSX 9 kb)
Supplementary Table 7
List of isolation conditions and classification of 23 Lachnospiraceae strains (XLSX 36 kb)
Rights and permissions
About this article
Cite this article
Chen, L., Wilson, J., Koenigsknecht, M. et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol 18, 541–551 (2017). https://doi.org/10.1038/ni.3690
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3690